by Ryan Pevey
TL;DR: I used Python to generate synthetic neurons inspired by the Brainbow technique. This post explores how simple modeling and iteration can create visually compelling, anatomically inspired neuronal art. Includes an interactive widget and gallery.
Let’s make some art!!!
I want to make something clear up front. I am not somebody who considers themselves artistic, but I do really enjoy making things visually appealing within the context of data and science communication. On top of that, I’m in the mood to make something that just kinda looks cool. So, if you’re not familiar with it, Brainbow is an iconic and classic fluorescent imaging technique in neuroscience that uses Cre/lox recombination to create stochastic expression of three or more fluorescent transgenes in a population of your target cells Livet, 2007. This stochastic integration of multiple fluorescent transgenes means that each adjacent neuron expresses a different permutation of each of the fluorescent tags thereby distinguishing itself from it’s neighbors and producing a visually stunning field of view in the resulting micrographs. In short, it’s a valuable scientific tool as well as a visual masterpiece.
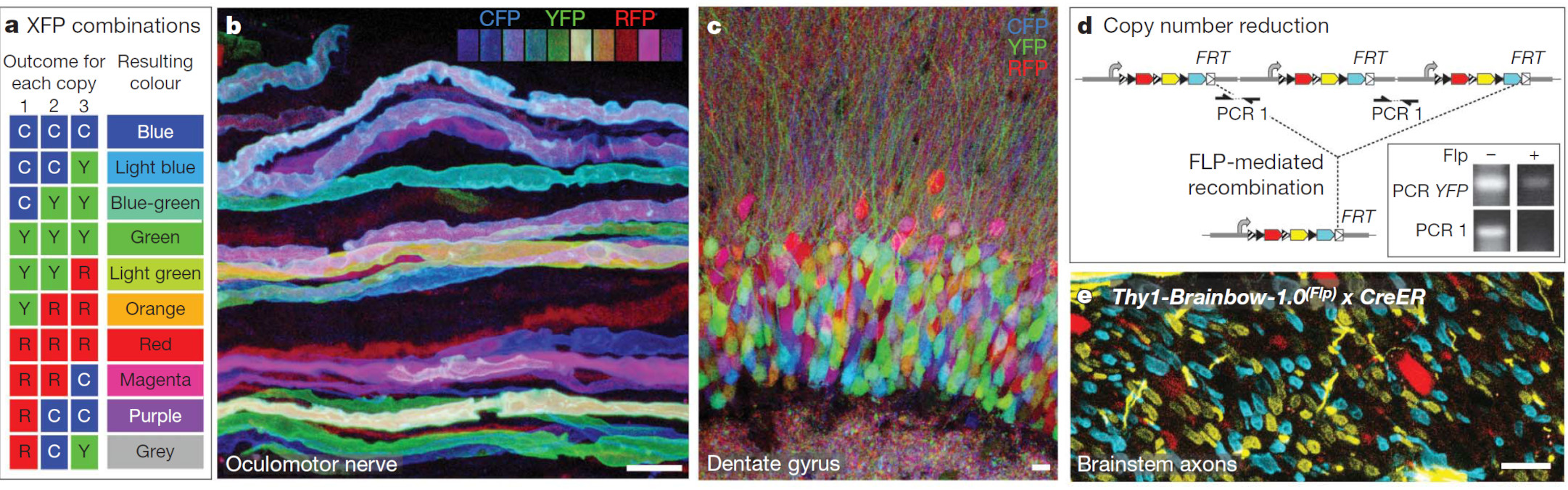
Fig. 1: Reprinted Fig. 4 from Livet, 2007. This figure demonstrates how Brainbow constructs enable diverse neuronal labeling through independent recombination of multiple tandem transgene copies, resulting in a spectrum of fluorescent expression. Example panels show multicolor labeling in oculomotor axons and dentate gyrus neurons, as well as how recombination can reduce transgene copy number or enforce mutually exclusive expression. Panel c. is what I’m aiming for, except for excitatory neurons in the neocortical plate.
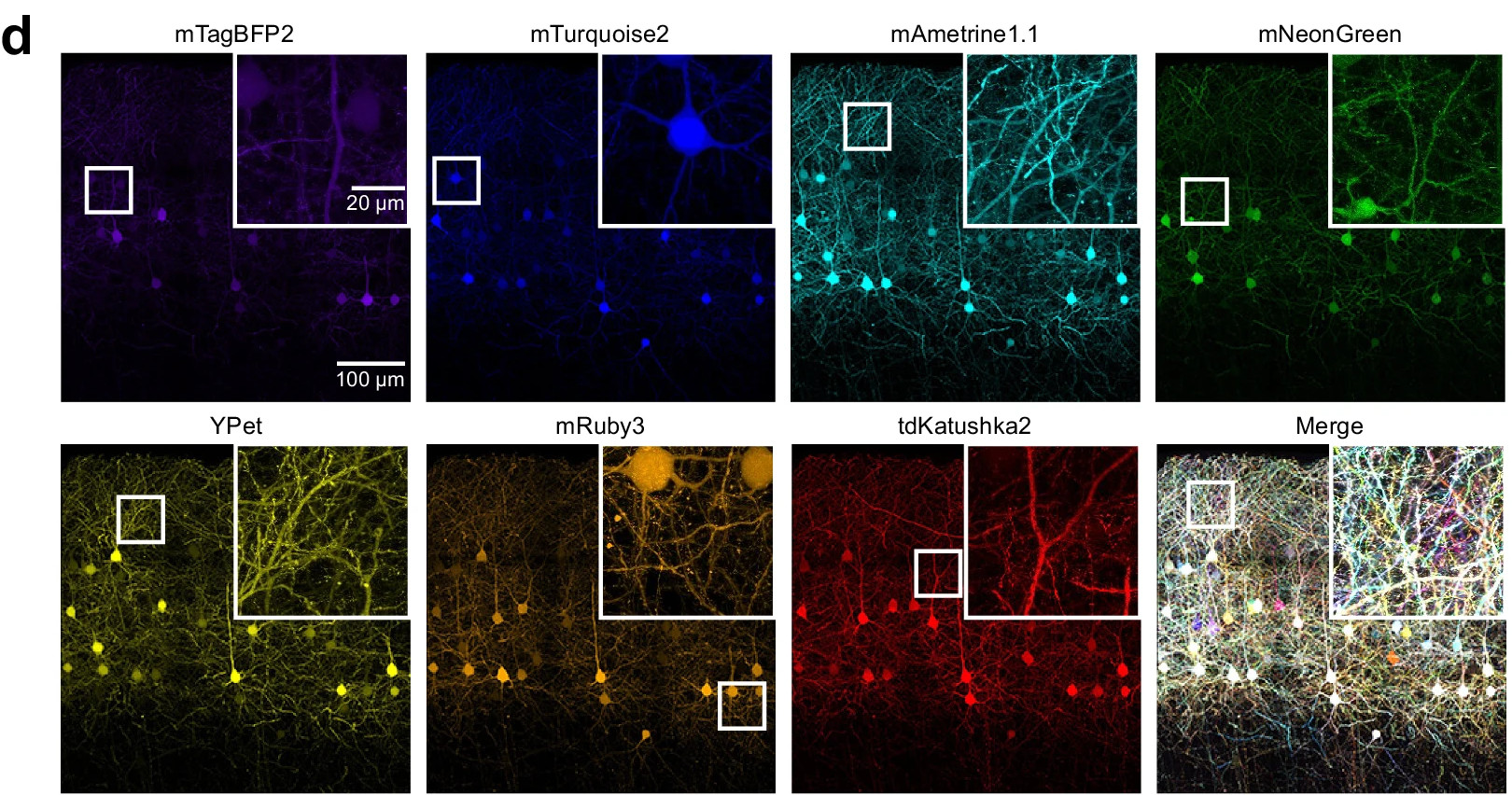
Fig. 2: Reprinted Fig. 1d from Leiwe, 2024. This figure shows layer 2/3 neurons in the mouse somatosensory cortex labeled using seven-color Brainbow based method, Tetbow, delivered by in utero electroporation. Fluorescent XFP signals were linearly unmixed and imaged in a 3D z-stack at postnatal day 28, revealing distinct color labeling of individual neurons.
These Brainbow images inspired me to try to recreate them visually for some digital art so I got to work trying to figure out a good way to do this. At first, I was going to create this using specialized neuronal modeling software, either by downloading annotated neuron anatomy models, or using research grade simplified models. I’ve toyed around with NEURON published by Yale University, Hines, 1997 before, so my initial impulse was to use that but that was years ago so I wanted to see what else was out there. I also tried a few other options (non-exhaustively): navis, NeuroM (Blue Brain Project), Treem, and Vedo for procedural generation control. Ultimately, and no offense meant to any of these fine packages, I realized that I was really just interested in making something visually interesting so I just ended up coding it in python with matplotlib. Those were all a fun aside and there’s some cool software out there nowadays, but there’s also something to be said for hard coding things yourself and building in an efficient pipeline with just the feature set that you need to get the job done. Anyway, let’s see how I did!
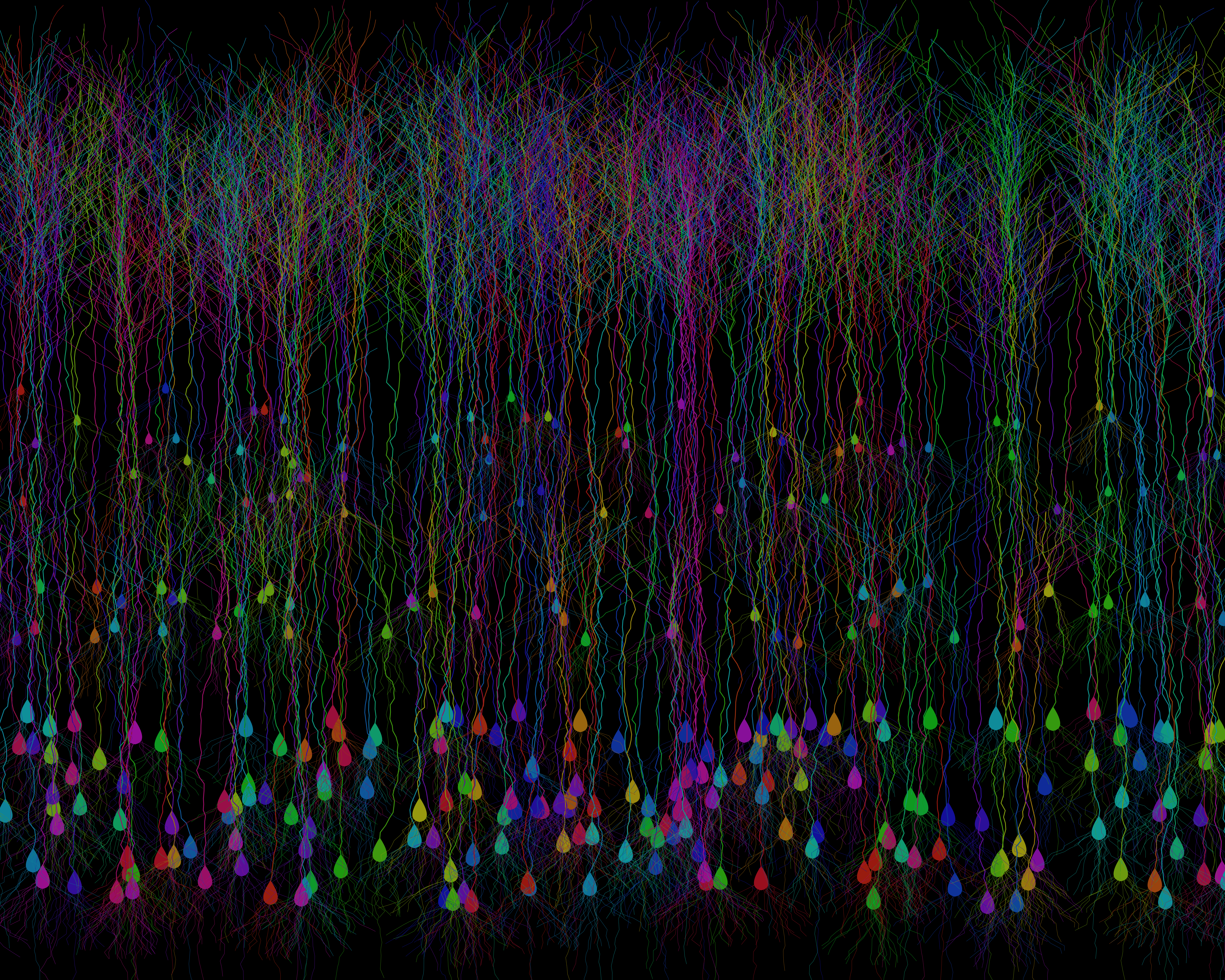
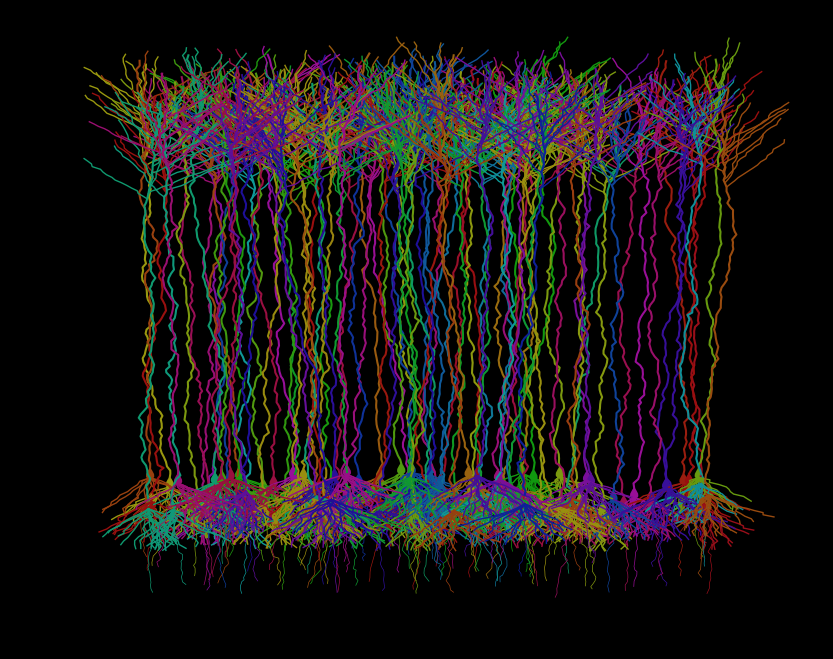
Fig. 3: A synthetic cortical field of Brainbow-inspired neurons, procedurally generated in Python. Each cell is uniquely colored to mimic the striking stochastic expression seen in fluorescent Brainbow labeling.
I gotta say, I’m pretty happy with this! True, it’s easy to nitpick this from a neuroanatomy standpoint. But for such a lightweight modeling of a generalized excitatory neuron anatomy, it’s produced a pretty solid cortical plate! It’s totally catching the vibe that I’m looking for. There’s a dense Layer 5/6 with large pyramidal neurons, a more diffuse Layer 2/3 with smaller pyramidal neurons, all of the apical dendrites are meeting up in layer 1 and there’s even a granule layer with more flattened anatomy. I’m pretty chuffed!
Let’s see how I got to this point.
I started off creating a function for basic neuronal anatomy for one neuron at a time. It creates a teardrop shaped soma, then grows an apical dendrite upwards for 100 step units from there with some random noise side to side and branches off in a series of terminal tufts out of the top 10% of dendrite. After that I generate a series of basal dendrites from the soma and an axon directly from the bottom of the soma. The color is assigned randomly in HSV color space1 and then plotted with matplotlib. I tweaked all of the parameters in the function until I got something that I was happy with. When I plot ten of them in the same plane evenly spaced from each other you can see the variability that I can get out of one function. Then I tweaked some of the parameters for better visibility when plotted in a dense field.
Image 1: A synthetic neuron procedurally generated in Python. Each iteration is uniquely colored to mimic the striking stochastic expression seen in fluorescent Brainbow labeling. Image 2: An array of ten neurons, evenly spaced in the field of view, generated with the same code. Image 3: An array of 100 neurons placed randomly with a uniform distribution on the horizontal plane in the field of view. Image 4: An array of 100 neurons modified for better visibility and placed randomly with a uniform distribution on the horizontal and vertical axes to create a band of soma across the field of view.
After that I created a parameterized version of the function so that I could create different populations of neurons on the fly and place them within defined bands within the field of view. To get a sense for the parameterized variables in the function take at look that the widget that I created below2. Try out changing the parameters and see what kind of basic morphologies that you can create. Again, it’s easy to nitpick the anatomy, but I’m just going for a general gestalt3 here, so that when I plot a few hundred of them at once with various tweaked parameters it looks about right, like a Monet. Can you create an L5/6 deep layer neuron, or an L2/3 superficial layer neuron? The L4 granule cell layer neurons are the biggest stretch, I cranked the apical dendrite length all the way down and made the tufts and basal dendrites extra “branchy”, but I think it’s close enough in the aggregate.

Fig. 3 again: A synthetic cortical field of Brainbow-inspired neurons, procedurally generated in Python. Each cell is uniquely colored to mimic the striking stochastic expression seen in fluorescent Brainbow labeling.
How about a viridis version?
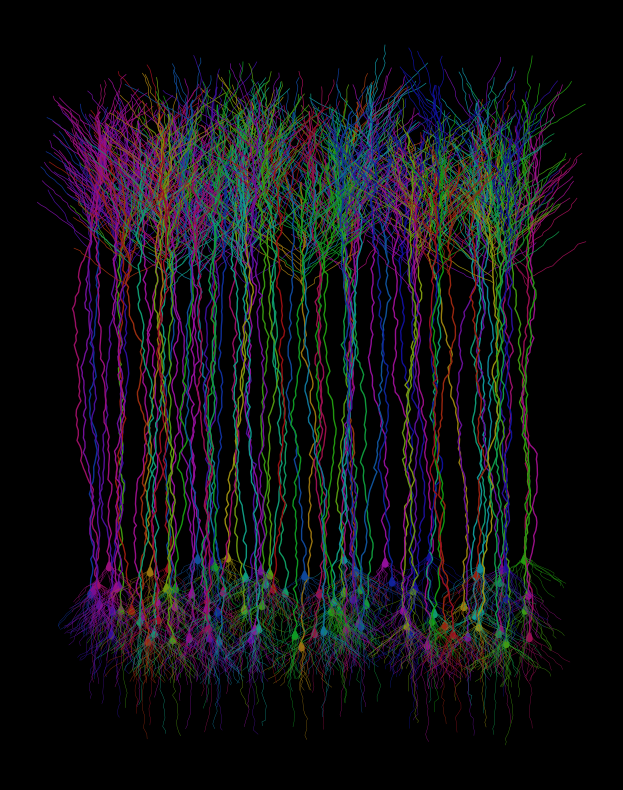
If you’ve read some of my other posts, you might know how big of a fan I am of the viridis color palette. So it’s pretty on-brand for me to make a viridis version of these images. Accessibility looks great on it!
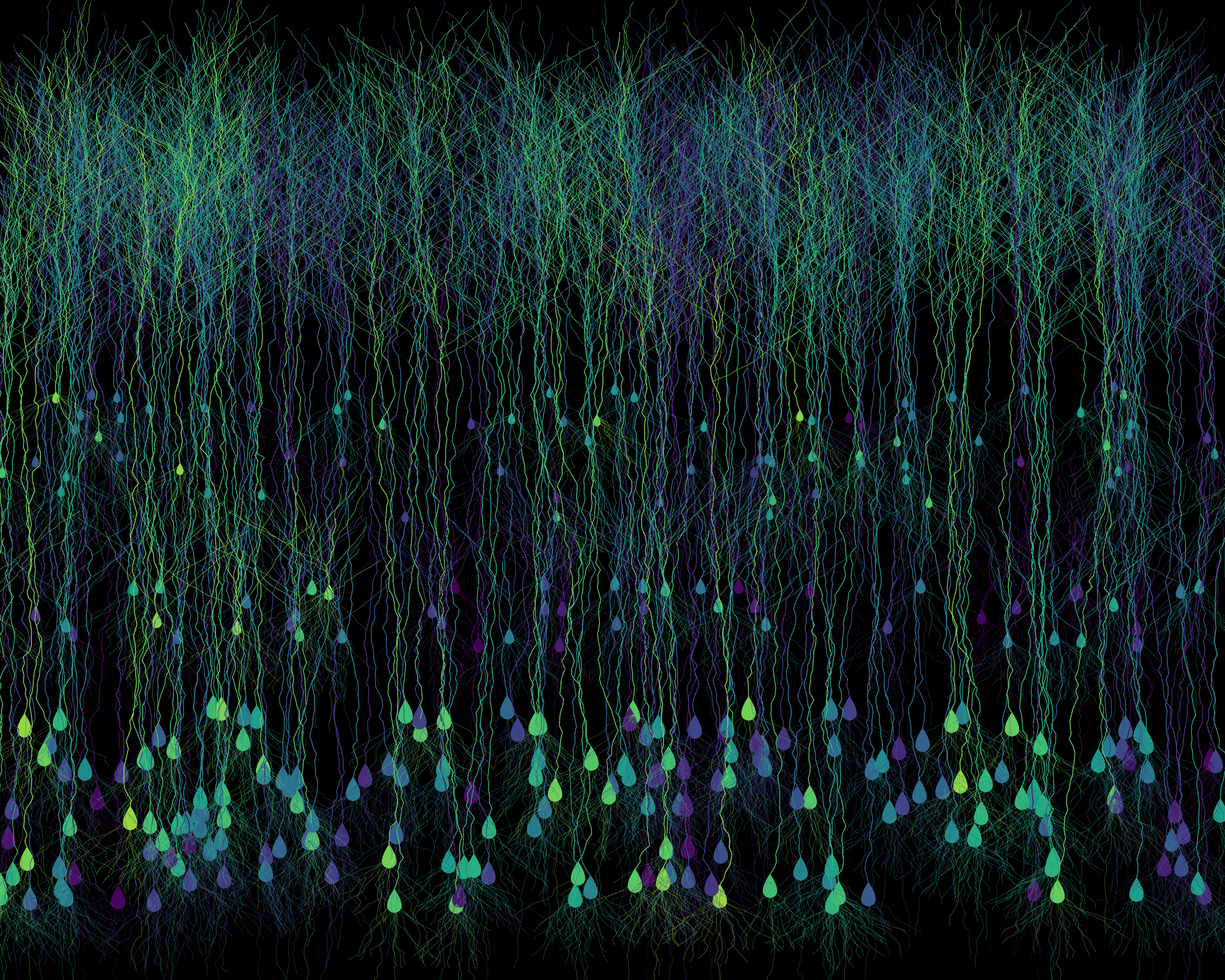
Fig. 4: A procedurally generated cortical field of digital neurons, each uniquely colorized using the Viridis palette, a perceptually uniform colorblind-friendly palette. The result is an artificial neural mosaic that’s more visually accessible than your average rainbow-washed microscopy.
Prints, Pixels & Possibly T-Shirts
Every so often, a visualization comes out looking a little too good to keep to myself. If you think so too and want to bring a little scientific art into your life, I’ve set up a Redbubble store where selected pieces are available. Visit my Redbubble shop. You can also browse the full gallery of curious creations here.
What I might add for the future
This was a pretty fun side project and I might do more of this in the future. I might, if I decide to revisit this make some more additions. Some extra things that I could add:
- Color generation using the 4-channel fluorescent staining to more faithfully recreate the brainbow staining pattern.
- More accurate neuronal anatomy by adding more customization of function used to procedurally generate these.
- Go all out by adding in the dozens of types of inhibitory neurons and interneurons!!!
Further Reading & References
- Original publication: Livet, J., Weissman, T., Kang, H. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007). 10.1038/nature06293
- Leiwe, M.N., Fujimoto, S., Baba, T. et al. Automated neuronal reconstruction with super-multicolour Tetbow labelling and threshold-based clustering of colour hues. Nat Commun 15, 5279 (2024). 10.1038/s41467-024-49455-y
- The NEURON simulation environment
- Hines ML, Carnevale NT. (1997). The NEURON simulation environment. Neural computation, 9(6), 1179-1209. doi:https://doi.org/10.1162/neco.1997.9.6.1179
- My gallery of images on evolio: evolio Gallery
- Color spaces and HSV
Scripts
You can find my github profile here: My GitHub profile.
Open data fuels open science.
Stay Connected
If you’re a student, researcher, or just a science enthusiast, I’d love to hear your thoughts. If you are a fan of inhibitory neurons or glial cells, what are your thoughts? Reach out or follow along via RSS for more deep dives into brain research, data storytelling, and big data reanalysis.
HSV (Hue, Saturation, Value) color space is one of several different color spaces that you can use in visualization and imaging. It was ideal for this application because the Hue is denoted by a single number representing an angle around the color wheel between 0 and 1. 0 is red, and as you increas to 1 you progress through the visibile spectrum through orange, yellow, green, blue, violet and magenta, then back around to red at 1. I set saturation at 0.9 and value at 0.6 to more closely match the colors in Brainbow staining, then converted hsv to rgb using the colorsys package. ↩
The code is recreated in javascript, p5.js to be specific, for computational efficiency for the webpage, but it generates a neuron the same way was the original python code. ↩
Gestalt is a German word meaning “form,” “shape,” or “configuration.” In psychology and related fields, it refers to a pattern or structure that is perceived as a unified whole, possessing qualities that cannot be understood merely by analyzing its individual components ↩


Leave a comment